maskfasta¶
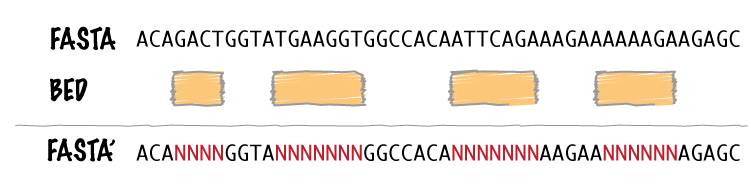
bedtools maskfasta masks sequences in a FASTA file based on intervals defined in a feature file. The
headers in the input FASTA file must exactly match the chromosome column in the feature file. This
may be useful for creating your own masked genome file based on custom annotations or for masking all
but your target regions when aligning sequence data from a targeted capture experiment.
Usage and option summary¶
Usage
$ bedtools maskfasta [OPTIONS] -fi <input FASTA> -bed <BED/GFF/VCF> -fo <output FASTA>
(or):
$ maskFastaFromBed [OPTIONS] -fi <input FASTA> -bed <BED/GFF/VCF> -fo <output FASTA>
Note
The input (-fi) and output (-fo) FASTA files must be different.
See also
| Option | Description |
|---|---|
| -soft | Soft-mask (that is, convert to lower-case bases) the FASTA sequence. By default, hard-masking (that is, conversion to Ns) is performed. |
| -mc | Replace masking character. That is, instead of masking with Ns, use another character. |
Default behavior¶
bedtools maskfasta will mask a FASTA file based on the intervals in a BED file. The newly masked FASTA file is written to the output FASTA file.
$ cat test.fa
>chr1
AAAAAAAACCCCCCCCCCCCCGCTACTGGGGGGGGGGGGGGGGGG
$ cat test.bed
chr1 5 10
$ bedtools maskfasta -fi test.fa -bed test.bed -fo test.fa.out
$ cat test.fa.out
>chr1
AAAAANNNNNCCCCCCCCCCGCTACTGGGGGGGGGGGGGGGGGG
-soft Soft-masking the FASTA file.¶
Using the -soft option, one can optionally “soft-mask” the FASTA file.
$ cat test.fa
>chr1
AAAAAAAACCCCCCCCCCCCCGCTACTGGGGGGGGGGGGGGGGGG
$ cat test.bed
chr1 5 10
$ bedtools maskfasta -fi test.fa -bed test.bed -fo test.fa.out -soft
$ cat test.fa.out
>chr1
AAAAAaaaccCCCCCCCCCCGCTACTGGGGGGGGGGGGGGGGGG
-mc Specify a masking character.¶
Using the -mc option, one can optionally choose a masking character to each base that will be masked by the BED file.
$ cat test.fa
>chr1
AAAAAAAACCCCCCCCCCCCCGCTACTGGGGGGGGGGGGGGGGGG
$ cat test.bed
chr1 5 10
$ bedtools maskfasta -fi test.fa -bed test.bed -fo test.fa.out -mc X
$ cat test.fa.out
>chr1
AAAAAXXXXXCCCCCCCCCCGCTACTGGGGGGGGGGGGGGGGGG
